Abstract
Background
The practise of prescribing Chronic Myeloid Leukaemia (CML) patients with indefinite tyrosine kinase inhibitors (TKIs) has gone uncontested, and the capacity of TKIs to eliminate the CML clone is uncertain. Although the vast majority of CML patients do respond to TKIs, nonetheless, resistance may develop either de novo or during treatment. TKI resistant pathways are commonly classified as BCR-ABL1-dependent or BCR-ABL1-independent. The causal factors in molecular evolution for this group of patients is still unknown. In this study, our goal was to explore the molecular mechanisms involved in patients who failed second-generation TKIs where we hope to identify potential genetic signatures and pathways that lead to TKI resistance.
Materials and Methods
CML patients who failed second generation TKI were identified and labeled as non-responders. Eight RNA samples were analysed for purity and processed using the Illumina Stranded Total RNA Prep, Ligation with Ribo-Zero Plus kit (Illumina, CA, USA) and were sequenced on Illumina Novaseq 6000 system (Illumina, CA, USA) system with an average of approximately 133 million passed paired reads, 100 bp in length, per sample. The paired-end reads were aligned to the human reference genome (version hg38/GRch38) using the Illumina DRAGEN (Dynamic Read Analysis for GENomics) Bio-IT Platform. The same system was used for gene-expression quantification, fusion gene detection, and variants analysis. Genes that demonstrate Log2 fold-change ≥3 and P≤0.01 were identified as differentially expressed genes (DEGs). The significantly enriched pathways from several pathway databases were filtered at a false-discovery rate (FDR) of <0.01.
Results
Variant analysis
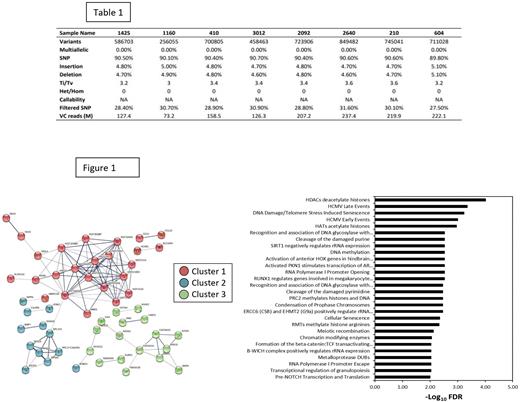
As shown in Table 1, an average of 628,935 ± 190,0862 variants were identified in the non-responder compared to 545,496 ± 58,000 variants in the responder. Out of the variants detected in the non-responder, 90.39% ± 0.30% was SNPs, 4.83% ± 0.15% insertions, and 4.78% ± 0.17% deletions. A total of 21,782 variants were found to be exclusively present in non-responder only.
Differentially Expressed Genes (DEGs)
Whole transcriptomic analysis identified a total of 201 differentially regulated genes (DEGs; Log2 fold-change ≥ 3 and P ≤ 0.01, Student's t-test), of which 87 genes were up-regulated and 114 genes down-regulated in the non-responder group.
Network and Gene Enrichment Analysis
The identified DEGs were subjected to network and gene enrichment analysis using STRING database. A total of 3 distinct clustered were identified, corresponding to histone acetylation/deacetylation (HDACs/HATs), cellular senescence, and RNA polymerase regulation as shown in the Figure 1.
Discussion & Conclusion
Techniques with enhanced sensitivity such as next-generation sequencing and the use of artificial intelligence techniques coupled with the development of mathematical modelling and computational prediction methods may reveal the underlying mechanism of drug resistance and facilitate the design of more effective treatment strategies in CML patients. This preliminary analysis found that individuals who failed second-generation TKIs expressed a potentially unique genetic signature. However, a larger sample size is necessary to verify these findings.
Disclosures
Selvaratnam:Apellis Pharmaceuticals: Consultancy. Syed Abd Kadir:MSD: Research Funding. Tan:Novartis: Honoraria, Research Funding; MSD: Honoraria; Amgen: Honoraria; BMS: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal